Synthentic Biological Sequence Design
MILA
The ability to accelerate the design of biological sequences can have a substantial impact on the progress of the medical field. The problem can be framed as a global optimization problem where the objective is an expensive black-box function such that we can query large batches restricted with a limitation of a low number of rounds. Bayesian Optimization is a principled method for tackling this problem. However, the astronomically large state space of biological sequences renders brute-force iterating over all possible sequences infeasible.
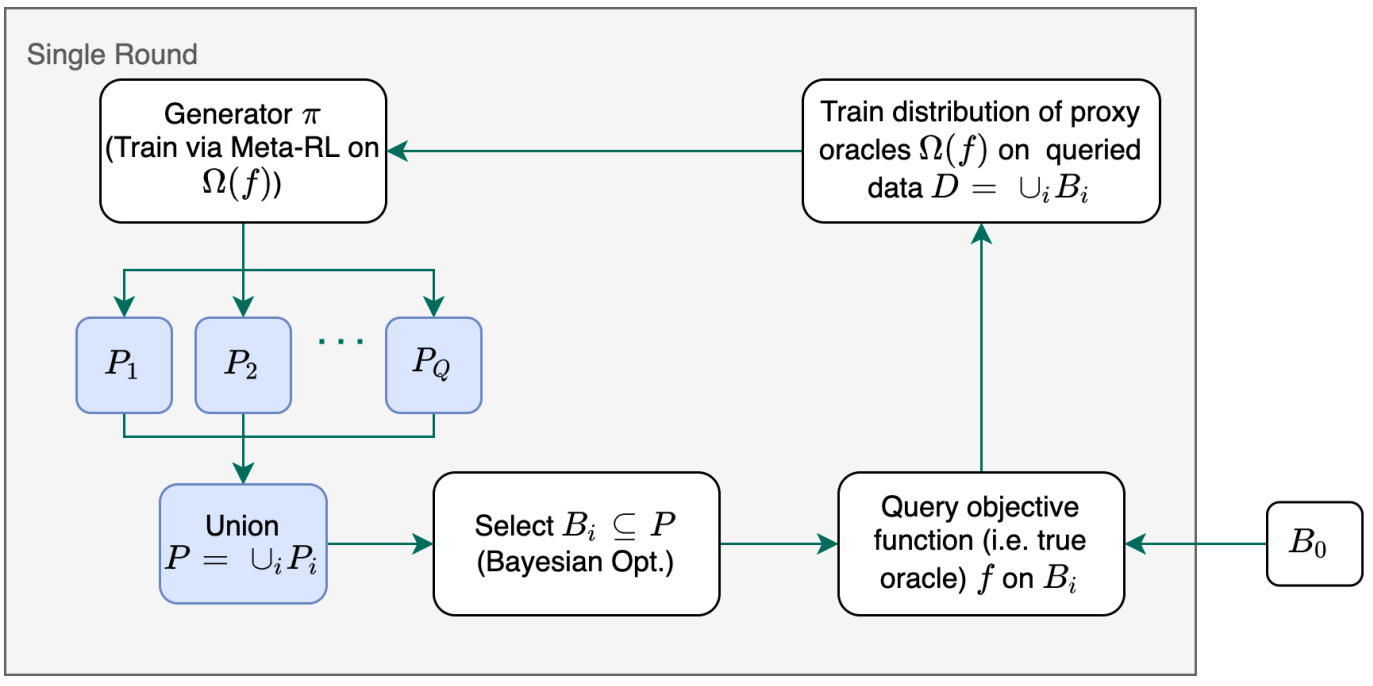
In this work, we propose MetaRLBO where we train an autoregressive generative model via Meta-Reinforcement Learning to propose promising sequences for selection via Bayesian Optimization. We pose this problem as that of finding an optimal policy over a distribution of MDPs induced by sampling subsets of the data acquired in the previous rounds. Our insilico experiments show that meta-learning over such ensembles provides robustness against reward misspecification and achieves better sample efficiency compared to existing state-of-the-art methods.
Publication:
[1] L. Feng, P. Nouri, A. Muni, Y. Bengio, P. Bacon.
Designing Biological Sequences via Meta-Reinforcement Learning
and Bayesian Optimization, 2022. (Under Review: 39th International
Conference on Machine Learning, ICML)